马闯等《Genomics, Proteomics & Bioinformatics》2021年
论文题目:Interactive Web-based Annotation of Plant MicroRNAs with iwa-miRNA
论文作者:Ting Zhang,Jingjing Zhai,Xiaorong Zhang,Lei Ling,Menghan Li,Shang Xie,Minggui Song,Chuang Ma
论文摘要:MicroRNAs (miRNAs) are important regulators of gene expression. The large-scale detection and profiling of miRNAs have accelerated with the development of high-throughput small RNA sequencing (sRNA-Seq) techniques and bioinformatics tools. However, generating high-quality comprehensive miRNA annotations remains challenging due to the intrinsic complexity of sRNA-Seq data and inherent limitations of existing miRNA predictions. Here, we present iwa-miRNA, a Galaxy-based framework that can facilitate miRNA annotation in plant species by combining computational analysis and manual curation. iwa-miRNA is specifically designed to generate a comprehensive list of miRNA candidates, bridging the gap between already annotated miRNAs provided by public miRNA databases and new predictions from sRNA-Seq datasets. It can also assist users in selecting promising miRNA candidates in an interactive mode, contributing to the accessibility and reproducibility of genome-wide miRNA annotation. iwa-miRNA is user-friendly and can be easily deployed as a web application for researchers without programming experience. With flexible, interactive, and easy-to-use features, iwa-miRNA is a valuable tool for the annotation of miRNAs in plant species with reference genomes. We illustrated the application of iwa-miRNA for miRNA annotation using data from plant species with varying genomic complexity. The source codes and web server of iwa-miRNA are freely accessible at: http://iwa-miRNA.omicstudio.cloud/.
论文链接:https://pubmed.ncbi.nlm.nih.gov/34332120/
iwa-miRNA:基于Web的可交互式植物microRNA注释平台
MicroRNA(miRNA)是一类长约21 nt的非编码小RNA分子,其通过在转录后水平调控基因的表达,在植物生长发育、逆境响应以及营养代谢等诸多生物过程中发挥着重要作用。近年来,高通量小RNA测序技术的快速发展和广泛应用极大地推进了植物miRNA的鉴定和功能研究。但是,高通量小RNA测序数据的复杂性和现有生物信息学分析方法的局限性使得在全基因组水平获得高质量的miRNA注释仍具有很大的挑战。近日,我院马闯教授团队基于Galaxy平台开发了一款新的miRNA注释软件iwa-miRNA,结合计算分析和人工筛选两种策略实现植物基因组中miRNA的高质量注释。iwa-miRNA内置了一系列的生物信息学分析流程,可自动聚合miRBase、PmiREN和sRNAanno等具有代表性的miRNA数据库的注释数据,挖掘高通量小RNA测序数据中的候选miRNA分子,刻画miRNA在序列、结构和表达等多个层面的特征信息,建立动态的Web交互界面实现高质量miRNA的人工辅助筛选,构建基于机器学习的计算生物学模型进行全基因组水平的miRNA精确预测。iwa-miRNA具有易用性强、实用性广和结果再现性好等优点,其所有程序代码及依赖的库文件均已集成在Docker镜像文件,可便捷地部署在Windows、Linux和Macintosh等多种操作系统上,进而搭建一个集多用户、多任务、多线程以及可回溯等功能于一体的植物miRNA注释服务器,实现基于Web或命令行的植物miRNA的大规模注释。iwa-miRNA的性能有效性已在拟南芥、玉米和中国春小麦上得到验证,其源码和Docker镜像文件可从https://github.com/cma2015/iwa-miRNA或http://iwa-miRNA.omicstudio.cloud网址获得。
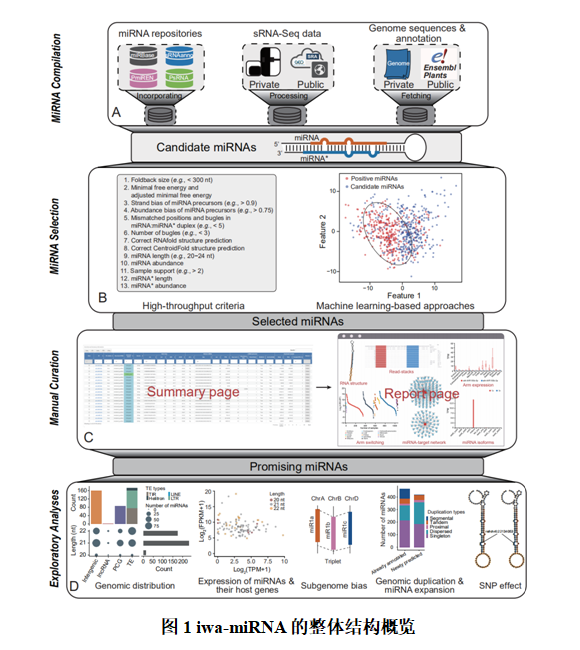
该项目由我院生物信息学中心马闯教授领导的科研创新团队完成,该团队的博士生张庭为该论文的第一作者,马闯教授为通讯作者。该研究工作得到了国家自然科学基金、国家青年人才项目、陕西省人才计划、中央高校基本科研业务费等项目的支持。西北农林科技大学高性能计算平台也为iwa-miRNA的研发提供了重要的计算资源。
